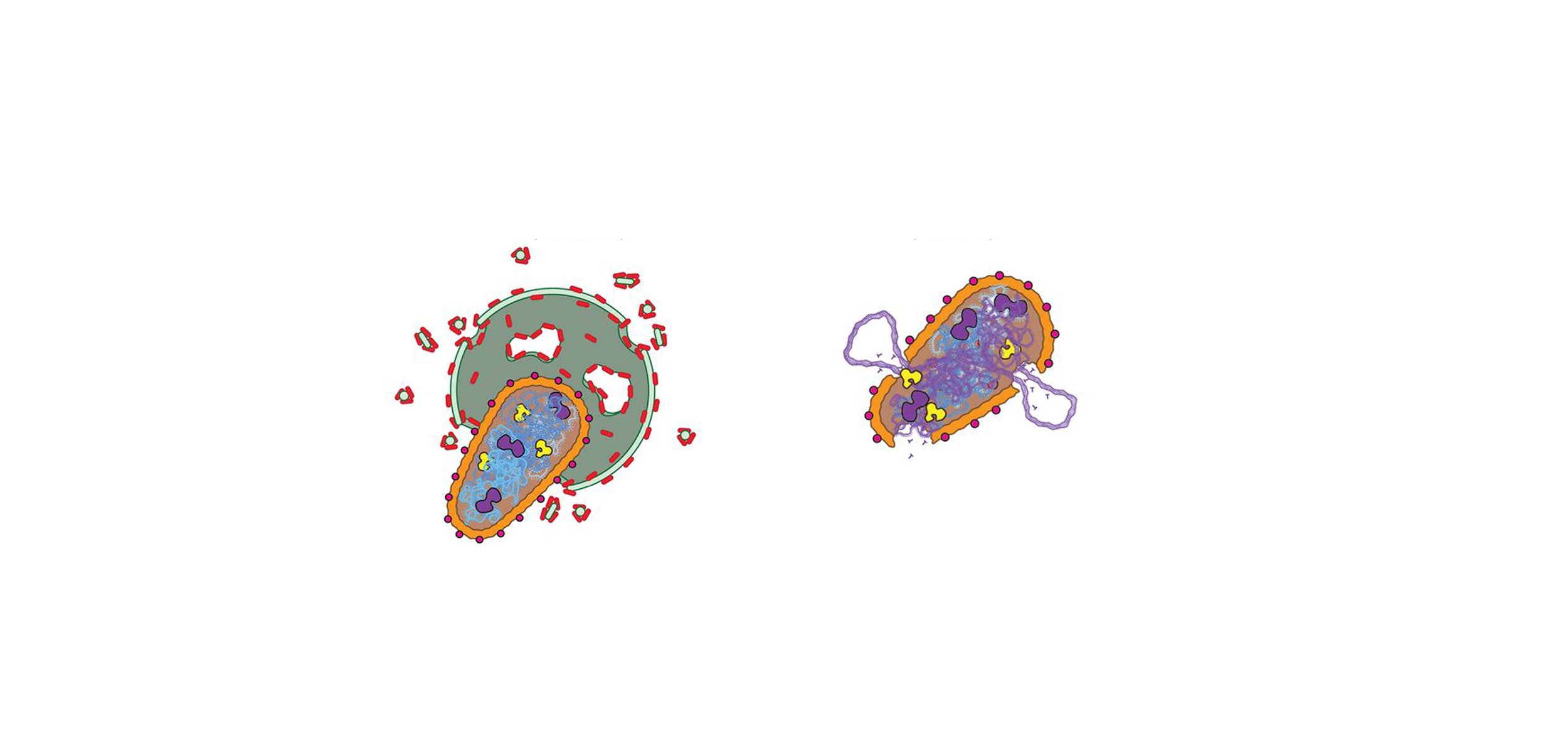
Publications
Thylakoid membrane remodeling by VIPP1 ESCRT-III-like filaments.
McCullough J, Sundquist WI. Nat Struct Mol Biol. 2025
Cell-free assays reveal that the HIV-1 capsid protects reverse transcripts from cGAS immune sensing.
Scott TM, Arnold LM, Powers JA, McCann DA, Rowe AB, Christensen DE, Pereira MJ, Zhou W, Torrez RM, Iwasa JH, Kranzusch PJ, Sundquist WI, Johnson JS. PLoS Pathog. 2025.
The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Paine EL, Whitby FG, Mackay DR, Ullman KS, Hill CP, Sundquist WI. Elife. 2023. PMCID: PMC10586806.
Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission.
Wenzel DM, Mackay DR, Skalicky JJ, Paine EL, Miller MS, Ullman KS, Sundquist WI. eLife. 2022. PMCID: PMC9477494
Recurrent evolution of an inhibitor of ESCRT-dependent virus budding and LINE-1 retrotransposition in primates.
Rheinemann L, Miller Downhour D, Davenport KA, McKeown AN, Sundquist WI, Elde NC.
Current Biology. 2022. PMCID: PMC9007875
Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro.
Swanstrom R, Sundquist WI. Viruses. 2021. PMCID: PMC8621278.
RetroCHMP3 blocks budding of enveloped viruses without blocking cytokinesis.
Rheinemann L, Downhour DM, Bredbenner K, Mercenne G, Davenport KA, Schmitt PT, Necessary CR, McCullough J, Schmitt AP, Simon SM, Sundquist WI, Elde NC. Cell. 2021. PMID: 34597582.
Identification of abscission checkpoint bodies as structures that regulate ESCRT factors to control abscission timing.
Williams LK, Mackay DR, Whitney MA, Couldwell GC, Sundquist WI, Ullman KS.
Elife. 2021. PMCID: PMC8437436.
AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
Rheinemann L, Thompson T, Mercenne G, Paine EL, Peterson FC, Volkman BF, Alam SL, Alian A, Sundquist WI.
J Biol Chem. 2021. PMCID: PMC8368996
Membrane Remodeling: ESCRT-III Filaments as Molecular Garrotes.
McCullough J, Sundquist WI. Curr Biol. 2020. PMID: 33290710.
Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro.
Christensen DE, Ganser-Pornillos BK, Johnson JS, Pornillos O, Sundquist WI.
Science. 2020. PMCID: PMC8022914.
Virus budding.
Sundquist WI, Rheinemann L. Encyclopedia of Virology. 2020 August.
Clinical targeting of HIV capsid protein with a long-acting small molecule.
Link JO, Rhee MS, Tse WC, Zheng J, Somoza JR, Rowe W, Begley R, Chiu A, Mulato A, Hansen D, Singer E, Tsai LK, Bam RA, Chou CH, Canales E, Brizgys G, Zhang JR, Li J, Graupe M, Morganelli P, Liu Q, Wu Q, Halcomb RL, Saito RD, Schroeder SD, Lazerwith SE, Bondy S, Jin D, Hung M, Novikov N, Liu X, Villaseñor AG, Cannizzaro CE, Hu EY, Anderson RL, Appleby TC, Lu B, Mwangi J, Liclican A, Niedziela-Majka A, Papalia GA, Wong MH, Leavitt SA, Xu Y, Koditek D, Stepan GJ, Yu H, Pagratis N, Clancy S, Ahmadyar S, Cai TZ, Sellers S, Wolckenhauer SA, Ling J, Callebaut C, Margot N, Ram RR, Liu YP, Hyland R, Sinclair GI, Ruane PJ, Crofoot GE, McDonald CK, Brainard DM, Lad L, Swaminathan S, Sundquist WI, Sakowicz R, Chester AE, Lee WE, Daar ES, Yant SR, Cihlar T. Nature. 2020. PMCID: PMC8188729.
Membrane constriction and thinning by sequential ESCRT-III polymerization.
Nguyen HC, Talledge N, McCullough J, Sharma A, Moss FR 3rd, Iwasa JH, Vershinin MD, Sundquist WI, Frost A.
Nat Struct Mol Biol. 2020. PMCID: PMC7343221.
Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
Han H, Schubert HL, McCullough J, Monroe N, Purdy MD, Yeager M, Sundquist WI, Hill CP.
J Biol Chem. 2020. PMCID: PMC6956536.
A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model.
Yant SR, Mulato A, Hansen D, Tse WC, Niedziela-Majka A, Zhang JR, Stepan GJ, Jin D, Wong MH, Perreira JM, Singer E, Papalia GA, Hu EY, Zheng J, Lu B, Schroeder SD, Chou K, Ahmadyar S, Liclican A, Yu H, Novikov N, Paoli E, Gonik D, Ram RR, Hung M, McDougall WM, Brass AL, Sundquist WI, Cihlar T, Link JO.
Nat Med. 2019. PMCID: PMC7396128.
Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Han H, Fulcher JM, Dandey VP, Iwasa JH, Sundquist WI, Kay MS, Shen PS, Hill CP. Elife. 2019 Jun 11;8. PMCID: PMC6602582.
Retrovirus Restriction by TRIM5α: RINGside at a Cage Fight.
Sundquist WI, Pornillos O. Cell Host Microbe. 2018. PMCID: PMC6464375.
Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes.
McCullough J, Frost A, Sundquist WI. Annu Rev Cell Dev Biol. 2018. PMCID: PMC6241870.
A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability.
Sadler JBA, Wenzel DM, Williams LK, Guindo-Martínez M, Alam SL, Mercader JM, Torrents D, Ullman KS, Sundquist WI, Martin-Serrano J. Proc Natl Acad Sci U S A. 2018. PMCID: PMC6156662.
The ESCRT-III proteins IST1 and CHMP1B assemble around nucleic acids.
Talledge N, McCullough J, Wenzel DM, Nguyen HC, Lalonde MS, Bajorek M, Skalicky J, Frost A, Sundquist WI. (2018)
bioRxiv
General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.
Wagner JM, Christensen DE, Bhattacharya A, Dawidziak DM, Roganowicz MD, Wan Y, Pumroy RA, Demeler B, Ivanov DN, Ganser-Pornillos BK, Sundquist WI, Pornillos O. J Virol. 2018. PMCID: PMC5790955.
The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Han H, Monroe N, Sundquist WI, Shen PS, Hill CP. Elife. 2017. PMCID: PMC5716660.
What's in a name?
Schaller MD, McDowell G, Porter A, Shippen D, Friedman KL, Gentry MS, Serio TR, Sundquist WI. Elife. 2017. PMCID: PMC5655148.
Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase.
Monroe N, Han H, Shen PS, Sundquist WI, Hill CP. Elife. 2017. PMCID: PMC5413351.
LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells.
Gu M, LaJoie D, Chen OS, von Appen A, Ladinsky MS, Redd MJ, Nikolova L, Bjorkman PJ, Sundquist WI, Ullman KS, Frost A. Proc Natl Acad Sci U S A. 2017. PMCID: PMC5358359.
Designed proteins induce the formation of nanocage-containing extracellular vesicles
Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, King NP, Sundquist WI. Nature. 2016. PMCID: PMC5729044.
Staff scientists in the workforce
Sundquist WI, Matthews CR. ASBMB Today. 2016.
Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids.
Li YL, Chandrasekaran V, Carter SD, Woodward CL, Christensen DE, Dryden KA, Pornillos O, Yeager M, Ganser-Pornillos BK, Jensen GJ, Sundquist WI. Elife. 2016. PMCID: PMC4936896.
Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α.
Wagner JM, Roganowicz MD, Skorupka K, Alam SL, Christensen D, Doss G, Wan Y, Frank GA, Ganser-Pornillos BK, Sundquist WI, Pornillos O. Elife. 2016. PMCID: PMC4936894.
Creating a robust research enterprise.
Sundquist WI, Corb B. ASBMB Today. 2016.
Structure and membrane remodeling activity of ESCRT-III helical polymers
McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, Hanson PI, Frost A. Science. 2015. PMCID: PMC4684769.
Toward a sustainable biomedical research enterprise: Finding consensus and implementing recommendations.
Pickett CL, Corb BW, Matthews CR, Sundquist WI, Berg JM. Proc Natl Acad Sci U S A. 2015. PMCID: PMC4568264.
TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription.
Fletcher AJ, Christensen DE, Nelson C, Tan CP, Schaller T, Lehner PJ, Sundquist WI, Towers GJ. EMBO J. 2015. PMCID: PMC4551353.
ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins.
Caballe A, Wenzel DM, Agromayor M, Alam SL, Skalicky JJ, Kloc M, Carlton JG, Labrador L, Sundquist WI, Martin-Serrano J. Elife. 2015. PMCID: PMC4475061.
Binding of Substrates to the Central Pore of the Vps4 ATPase Is Autoinhibited by the Microtubule Interacting and Trafficking (MIT) Domain and Activated by MIT Interacting Motifs (MIMs).
Han H, Monroe N, Votteler J, Shakya B, Sundquist WI, Hill CP. J Biol Chem. 2015. PMCID: PMC4505595.
Angiomotin functions in HIV-1 assembly and budding.
Mercenne G, Alam SL, Arii J, Lalonde MS, Sundquist WI. Elife. 2015. PMCID: PMC4337731.
Putting a finger in the ring.
McCullough J, Sundquist WI. Nat Struct Mol Biol. 2014. PMID: 25469842.
The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer.
Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O. Proc Natl Acad Sci U S A. 2014. PMCID: PMC3932864.
The oligomeric state of the active Vps4 AAA ATPase.
Monroe N, Han H, Gonciarz MD, Eckert DM, Karren MA, Whitby FG, Sundquist WI, Hill CP. J Mol Biol. 2014. PMCID: PMC3919030.
ESCRT requirements for EIAV budding.
Sandrin V, Sundquist WI. Retrovirology. 2013. PMCID: PMC3907061.
Discovery of novel small-molecule HIV-1 replication inhibitors that stabilize capsid complexes.
Lamorte L, Titolo S, Lemke CT, Goudreau N, Mercier JF, Wardrop E, Shah VB, von Schwedler UK, Langelier C, Banik SS, Aiken C, Sundquist WI, Mason SW. Antimicrob Agents Chemother. 2013. PMCID: PMC3811413.
Virus budding and the ESCRT pathway.
Votteler J, Sundquist WI. Cell Host Microbe. 2013. PMCID: PMC3819203.
Macromolecular assemblies.
Rey FA, Sundquist WI. Curr Opin Struct Biol. 2013. PMID: 23663961.
Membrane fission reactions of the mammalian ESCRT pathway.
McCullough J, Colf LA, Sundquist WI. Annu Rev Biochem. 2013. PMCID: PMC4047973.
Building a super elongation complex for HIV.
Hill CP, Sundquist WI. Elife. 2013. PMCID: PMC3592195.
ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding.
Effantin G, Dordor A, Sandrin V, Martinelli N, Sundquist WI, Schoehn G, Weissenhorn W. Cell Microbiol. 2013. PMCID: PMC3783020.
Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport.
Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, Uter NT, Bajorek M, Myszka DG, Sundquist WI. J Biol Chem. 2012. PMCID: PMC3527974.
ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding.
Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR. Dev Cell. 2012. PMCID: PMC3522770.
How HIV finds the door.
Lalonde MS, Sundquist WI. Proc Natl Acad Sci U S A. 2012. PMCID: PMC3503163.
Attenuated protein expression vectors for use in siRNA rescue experiments.
Morita E, Arii J, Christensen D, Votteler J, Sundquist WI. Biotechniques. 2012. PMCID: PMC3759224.
HIV-1 assembly, budding, and maturation.
Sundquist WI, Kräusslich HG. Cold Spring Harb Perspect Med. 2012. PMCID: PMC3385941.
Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein.
Lemke CT, Titolo S, von Schwedler U, Goudreau N, Mercier JF, Wardrop E, Faucher AM, Coulombe R, Banik SS, Fader L, Gagnon A, Kawai SH, Rancourt J, Tremblay M, Yoakim C, Simoneau B, Archambault J, Sundquist WI, Mason SW. J Virol. 2012. PMCID: PMC3393593.
Assemble, replicate, remodel and evade.
Sundquist WI, Krug RM. Curr Opin Virol. 2012. PMID: 22482706.
Global landscape of HIV-human protein complexes.
Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. Nature. 2011. PMCID: PMC3310911.
Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding.
Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. PLoS One. 2011. PMCID: PMC3230590.
Activation of the retroviral budding factor ALIX.
Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. J Virol. 2011. PMCID: PMC3165844.
Macromolecular assemblages.
Rey FA, Sundquist WI. Curr Opin Struct Biol. 2011. PMID: 21419622.
ESCRT-III protein requirements for HIV-1 budding.
Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. Cell Host Microbe. 2011. PMCID: PMC3070458.
Hexagonal assembly of a restricting TRIM5alpha protein.
Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Proc Natl Acad Sci U S A. 2011. PMCID: PMC3021009.
Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1.
hai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. J Virol. 2011. PMCID: PMC3014167.
Commentary on "The Protein Network of HIV Budding" .
Sundquist WI. HIV: 30 years in 30 weeks; Most influential scientific papers on HIV published from 1981 to 2011. PubMed. 2011.
Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations.
Schubert HL, Zhai Q, Sandrin V, Eckert DM, Garcia-Maya M, Saul L, Sundquist WI, Steiner RA, Hill CP. Proc Natl Acad Sci U S A. 2010. PMCID: PMC2964199.
Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance.
Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Proc Natl Acad Sci U S A. 2010. PMCID: PMC2919903.
Structural basis for ESCRT-III protein autoinhibition.
Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Nat Struct Mol Biol. 2009 Jul;16(7):754-62. doi: 10.1038/nsmb.1621. Epub 2000. PMCID: PMC2712734.
X-ray structures of the hexameric building block of the HIV capsid.
Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. Cell. 2009. PMCID: PMC2840706.
Biochemical analyses of human IST1 and its function in cytokinesis.
Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Mol Biol Cell. 2009. PMCID: PMC2649257.
Biochemical and structural studies of yeast Vps4 oligomerization.
Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. J Mol Biol. 2008. PMCID: PMC2632936.
Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication.
Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, Sundquist WI. J Virol. 2008. PMCID: PMC2583683.
Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding.
Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Dev Cell. 2008. PMCID: PMC2586299.
ALIX-CHMP4 interactions in the human ESCRT pathway.
cCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. Proc Natl Acad Sci U S A. 2008. PMCID: PMC2409388.
NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains.
hung HY, Morita E, von Schwedler U, Müller B, Kräusslich HG, Sundquist WI. J Virol. 2008. PMCID: PMC2346761.
The structural biology of HIV assembly.
Ganser-Pornillos BK, Yeager M, Sundquist WI. Curr Opin Struct Biol. 2008. PMCID: PMC2819415.
Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p.
Yu Z, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. J Mol Biol. 2008. PMCID: PMC2279015.
Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV.
Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Nat Struct Mol Biol. 2008. PMID: 18066081.
Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein.
Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. J Mol Biol. 2007. PMCID: PMC2066180.
ESCRT-III recognition by VPS4 ATPases.
Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. Nature. 2007. PMID: 17928862.
How to assemble a capsid.
Sundquist WI, Hill CP. Cell. 2007. PMID: 17923081.
Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis.
Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. EMBO J. 2007. PMCID: PMC2230844.
An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein.
Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. Cell Host Microbe. 2007. PMCID: PMC3793644.
Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding.
Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. Cell Host Microbe. 2007. PMCID: PMC2034311.
The molecular mechanism of hepcidin-mediated ferroportin down-regulation.
De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. Mol Biol Cell. 2007. PMCID: PMC1924807.
Molecular characterization of the Ran-binding zinc finger domain of Nup153.
Higa MM, Alam SL, Sundquist WI, Ullman KS. J Biol Chem. 2007. PMID: 17426026
Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells.
Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. EMBO J. 2007. PMCID: PMC1852790.
Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding.
Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Cell. 2007. PMID: 17350572.
Macromolecular assemblages: focusing in on complexity.
Nogales E, Sundquist WI. Curr Opin Struct Biol. 2007.
The Polycomb-associated protein Rybp is a ubiquitin binding protein.
Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. FEBS Lett. 2006. PMID: 17070805.
Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain.
Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI. Nat Struct Mol Biol. 2006. PMID: 17057716.
Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release.
Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. J Virol. 2006. PMCID: PMC1617254.
Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1-278) and CA(N)(133-278).
Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Biochemistry. 2006. PMID: 16981686.
Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor.
Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Proc Natl Acad Sci U S A. 2006. PMCID: PMC1459386.
Two new structures of Ub-receptor complexes. U2.
Alam SL, Sundquist WI. Nat Struct Mol Biol. 2006. PMID: 16518384.
Structural and mechanistic studies of VPS4 proteins.
Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. EMBO J. 2005. PMCID: PMC1276703.
Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A.
Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Proc Natl Acad Sci U S A. 2005. PMCID: PMC1236530.
The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells.
Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J. J Biol Chem. 2005. PMID: 15644320.
Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus.
Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. J Virol. 2005. PMCID: PMC548478.
Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography
Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Mol Biol. 2005. PMCID: PMC6608732.
The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding.
Stuchell MD, Garrus JE, Müller B, Stray KM, Ghaffarian S, McKinnon R, Kräusslich HG, Morham SG, Sundquist WI. J Biol Chem. 2004. PMID: 15218037.
Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid.
Hatziioannou T, Cowan S, Von Schwedler UK, Sundquist WI, Bieniasz PD. J Virol. 2004. PMCID: PMC415825.
Ubiquitin interactions of NZF zinc fingers.
Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. EMBO J. 2004 . PMCID: PMC391057.
Ubiquitin recognition by the human TSG101 protein.
Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Mol Cell. 2004. PMID: 15053872.
Assembly properties of the human immunodeficiency virus type 1 CA protein.
Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. J Virol. 2004. PMCID: PMC369201.
Retrovirus budding.
Morita E, Sundquist WI. Annu Rev Cell Dev Biol. 2004. PubMed PMID: 15473846.
The protein network of HIV budding.
von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Kräusslich HG, Kaplan J, Morham SG, Sundquist WI. Cell. 2003. PMID: 14505570.
HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein.
Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. J Cell Biol. 2003. PMCID: PMC2172688.
Structure and ubiquitin binding of the ubiquitin-interacting motif.
Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP. J Biol Chem. 2003. PMID: 12750381.
Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly.
Ganser BK, Cheng A, Sundquist WI, Yeager M. EMBO J. 2003. PMCID: PMC162131.
Structural insights into the catalytic mechanism of cyclophilin A.
Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Nat Struct Biol. 2003. PMID: 12730686.
Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6(Gag).
Jenkins Y, Pornillos O, Rich RL, Myszka DG, Sundquist WI, Malim MH. J Virol. 2001. PMCID: PMC114634.
Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding.
Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI. Cell. 2001. PMID: 11595185.
Structural Models for the HIV-1 Capsid.
Li S, Ganser B, Hill CP, Finch JT, Sundquist WI. Virus Res. 2001.
Image reconstructions of helical assemblies of the HIV-1 CA protein.
Li S, Hill CP, Sundquist WI, Finch JT. Nature. 2000. PMID: 11014200.
Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Acta Crystallogr D Biol Crystallogr. 1999. PMID: 10089398
Assembly and analysis of conical models for the HIV-1 core.
Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Science. 1999. PMID: 9872746.
Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly.
von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. EMBO J. 1998. PMCID: PMC1170503.
Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Vajdos FF, Yoo S, Houseweart M, Sundquist WI, Hill CP. Protein Sci. 1997. PMCID: PMC2143596.
Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Science. 1997. PubMed PMID: 9346481.
Molecular recognition in the HIV-1 capsid/cyclophilin A complex.
Yoo S, Myszka DG, Yeh C, McMurray M, Hill CP, Sundquist WI. J Mol Biol. 1997. PubMed PMID: 9223641.
Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid.
Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Cell. 1996. PMID: 8980234.
Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses.
Christensen AM, Massiah MA, Turner BG, Sundquist WI, Summers MF. J Mol Biol. 1996.
Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: evidence for conformational changes during viral assembly.
Massiah MA, Worthylake D, Christensen AM, Sundquist WI, Hill CP, Summers MF. Protein Sci. 1996. PMCID: PMC2143307.
Structure of the amino-terminal core domain of the HIV-1 capsid protein.
Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Science. 1996. PMID: 8662505.
Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly.
Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Proc Natl Acad Sci U S A. 1996. PMCID: PMC39768.
Tattle Tales.
Sundquist WI. Nat Struct Biol. 1996. PMID: 8548459.
Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein.
Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. J Mol Biol. 1994. PMID: 7966331.
Synthesis, Characterization, DNA Binding Properties, and Solution Thermochromism of Platinum (II) Complexes of the Ethidium Cation: Altered Regioselectivity in a DNA-Promoted Reaction.
Ren T, Bancroft DP, Sundquist WI, Keck M, Masschelein A, Lippard SJ. Ren T, Bancroft DP, Sundquist WI, Keck M, Masschelein A, Lippar SJ (1993). J Am Chem Soc.
Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA.
Sundquist WI, Heaphy S. Proc Natl Acad Sci U S A. 1993. PMCID: PMC46306.
Mechanistic studies of a novel class of trisubstituted platinum(II) antitumor agents..
Hollis LS, Sundquist WI, Burstyn JN, Heiger-Bernays WJ, Bellon SF, Ahmed KJ, Amundsen AR, Stern EW, Lippard SJ. Cancer Res. 1991. PMID: 2004370.
The Structures of Telomeric DNA.
Sundquist WI. Nucleic Acids Mol Biol. 1991.
The Coordination Chemistry of Platinum Anticancer Drugs and Related Compounds with DNA.
Sundquist WI, Lippard SJ. Coord Chem Rev. 1990.
Synthesis, Characterization, and Biological Activity of cis-Diammineplatinum(II) Complexes of the DNA Intercalators 9-Aminoacridine and Chloroquine.
Sundquist WI, Bancroft DP, Lippard SJ. J Am Chem Soc. 1990.
Synthesis, Characterization, and DNA Binding Properties of (1,2-Diaminoethane)-platinum(II) Complexes Linked to the DNA Intercalator Acridine Orange by Trimethylene and Hexamethylene Chains.
Bowler BE, Ahmed KJ, Sundquist WI, Hollis LS, Whang EE, Lippard SJ. J Am Chem Soc. 1989.
DNA promotes the reaction of cis-diamminedichloroplatinum(II) with the exocyclic amino groups of ethidium bromide.
Sundquist WI, Bancroft DP, Chassot L, Lippard SJ. J Am Chem Soc. 1988.
Monoclonal antibodies to DNA modified with cis- or trans-diamminedichloroplatinum(II).
Sundquist WI, Lippard SJ, Stollar BD. Proc Natl Acad Sci U S A. 1987. PMCID: PMC299514.
Solvolysis reactions of cis- and trans-diamminedichloroplatinum(II) in dimethyl sulfoxide. Structural characterization and DNA binding of trans-bis(ammine)chloro(DMSO)platinum(1+).
Sundquist WI, Ahmed KJ, Hollis LS, Lippard SJ. Inorg Chem. 1987
Binding of cis- and trans-diamminedichloroplatinum(II) to deoxyribonucleic acid exposes nucleosides as measured immunochemically with anti-nucleoside antibodies.
Sundquist WI, Lippard SJ, Stollar BD. Biochemistry. 1986. PMID: 3707890.
Chemical and enzymatic biotin-labeling of oligodeoxyribonucleotides.
Kempe T, Sundquist WI, Chow F, Hu SL. Nucleic Acids Res. 1985. PMCID: PMC340973.
Multiple-copy genes: production and modification of monomeric peptides from large multimeric fusion proteins.
Kempe T, Kent SB, Chow F, Peterson SM, Sundquist WI, L'Italien JJ, Harbrecht D, Plunkett D, DeLorbe WJ. Gene. 1985. PMID: 2419204.
Selective 2'-benzoylation at the cis 2',3'-diols of protected ribonucleosides. New solid phase synthesis of RNA and DNA-RNA mixtures.
Kempe T, Chow F, Sundquist WI, Nardi TJ, Paulson B, Peterson SM. Nucleic Acids Res. 1982. PMCID: PMC326958.
